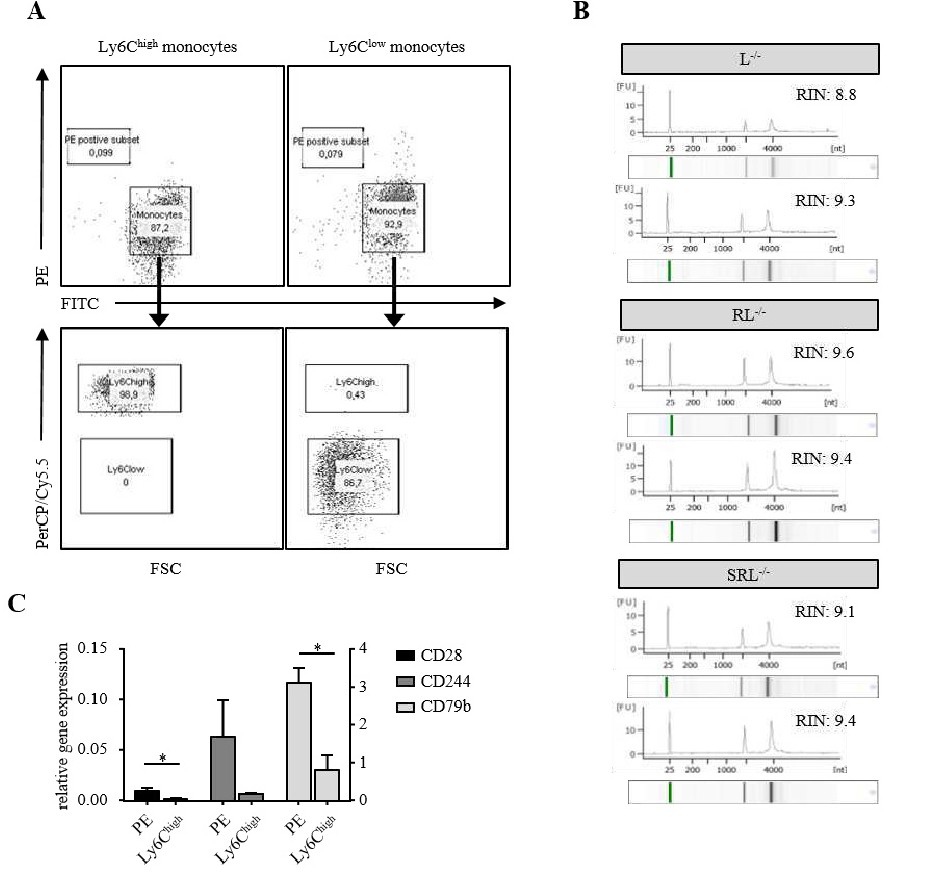
Fig. 2. Quality control of the sorted Ly6Chigh and Ly6Clow monocyte subsets and the mRNA isolated from these cells. (A) Re-sort of Ly6Chigh and Ly6Clow monocyte subsets by FACS exhibited a purity of 98.9% and 86.7% respectively. (B) Validation of RNA integrity that was isolated from Ly6Clow monocyte subsets from L-/-, RL-/- and SRL-/- mice was performed with an Agilent 2000 Bioanalyzer system. All RNA samples that were included in the microarray analysis were validated, but only two exemplary electropherograms and the corresponding RIN numbers of RNA are shown. (C) Validation of markers specific for B cells (CD79b), T cells (CD28) and NK cells (CD244) in Ly6Chigh monocyte subsets by real-time quantitative PCR (qPCR) in order to determine if the sorted cells originate from monocytes. The sorted PE-positive cell fraction served as positive control. Data are shown as mean ▒ SEM of 3-4 mice per group from duplicates of two individual experiments. Significance was calculated by multiple t test and statistical significance was determined using the Holm-Sidak method with alpha=5%.